Scientific Review
This web page was produced as an assignment for Gen677 at UW-Madison Spring 2009
SIRT6 links histone H3 lysine 9 deacetylation to NF-kB-dependent gene expression and organismal life span.
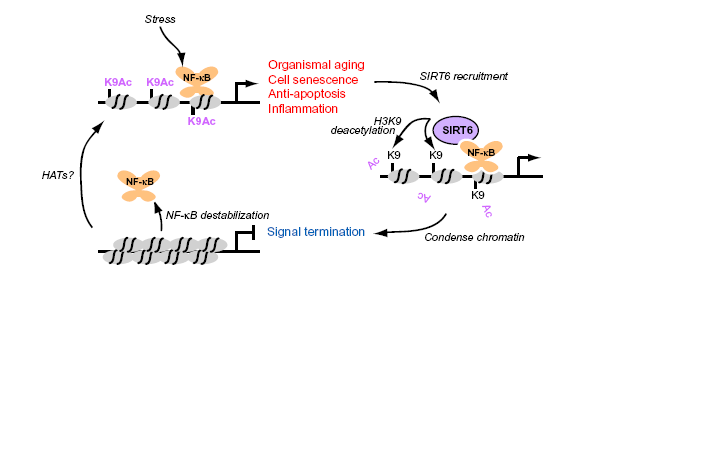
In this research article "SIRT6 links histone H3 lysine 9 deacetylation to NF-kB dependent gene expression and organismal life span" Kawahara et al show for the first time that SIRT6 deacetylates lysine 9 of histone H3 which leads to decreased expression of NF-kB genes. This is a novel discovery and brings insight into why SIRT6 knockout mice have shorter life spans.
Kawahara et al first determined that SIRT6 and the DNA binding domain of NF-kB where associating with each other through IPs (figure1). This demonstrates that SIRT6 associates with RELA lead them to look to see if SIRT6 is brought to the promoter of REL genes. To see if this where true they used ChIP, which allows investigators to determine DNA binding site of proteins, they found that indeed SIRT6 would associate at RELA promoters. Then to be sure that SIRT6 locating to promoters of RELA genes was due only to SIRT6 association with RELA they performed knock down experiments of RELA with shRNA and saw SIRT6 didn't locate to those promoters. The finding that SIRT6 is targeted to RELA promoters brought about the possibility that SIRT6 may influence expression of these genes. To test this possibility they used endogenous, overexpressed and shRNA knock down SIRT6. And coupling these different states with reporter genes and RT-PCR they determined that SIRT6 knockout lead to increased expression of RELA genes indicating that SIRT6 suppresses expression of RELA genes (figure2). And the genes that SIRT6 was shown to repress where genes associated with increased expression during aging (figure2). Then the authors used the recent knowledge that SIRT6 specifically locates at histones with H3K9 and deacetylates these histones to control gene expression to see if SIRT6 is required for deacetylating H3K9 histones of NF-kB genes. Again using ChIP of cells stimulated with TNF, an activator of NF-kB, to see which NF-kB genes had histone of H3K9 that where acetylated. Then using mouse cells that were missing SIRT6, it was observed that these cells had similar histone H3K9 acetylation as TNF stimulated cells. This allowed them to conclude that SIRT6 is recruited to promoters of NF-kB genes where there are H3K9 acetylations. And using similar methods as above Kawahara determined that the RELA subunit of NF-kB of SIRT6 knockout cells had similar occupancy at NF-kB gene as induced NF-kB. This definitively confirms that SIRT6 functions to destabilizes NF-kB at its promoters by deacetylating H3K9, thereby silencing these genes.
Kawahara next looked to see if the functions of NF-kB where effected by loss of SIRT6 and if this loss of SIRT6 brings about these effects through NF-kB. So looking at SIRT6 knockout cells they observed similar antiapoptotic and cell senescence by NF-kB through SIRT6 loss. This showed Kawahara that SIRT6 is critical in controlling apoptosis and cell senescence via NF-kB. Then to explore the effects that SIRT6 deficiency has on NF-kB and other transcription factors, they used SIRT6 knockout mice and wild type mice which were then subjected to competitive hybridization with microarrays. From this they were able to identify 4 transcription factors (NF-kB, CEBP, LFA1, and TEF) that where induced in SIRT6 knockout mice compared to wildtype mice. What was particular interesting about these transcription factors is they have been associated with changes in aging tissues. This paper so clearly demonstrated that SIRT6 is a critical component in controlling gene expression of NF-kB genes. And with the loss of SIRT6 there is an increased aging phenotype that is due to SIRT6 not deacetylating H3K9 of histones at key NF-kB genes.
This paper does a great job of demonstrating that SIRT6 is critical in regulating NF-kB genes through its deacetylation of H3K9 (as seen in the schematic drawing in the top of the page). This is so elegantly demonstrated through the use of ChIPs and IPs, to show in vivo that 1) SIRT6 associates with NF-kB subunit 2) that SIRT6 associates at sites where NF-kB binds its promoters. Then using SIRT6 knockout mice the investigators are able to use a very in vivo system to see similar results with cells. Some things I would have liked to see where if SIRT6 overexpressing mice had less NF-kB occupying promoter regions of their gene targets and if SIRT6 overexpressing mice have a longer life span. While these things I think would be interesting to see are beyond the scope of this paper. One critical concern I have is that SIRT1 was also able to bind to associate with RELA. So maybe some of these observations could be due to SIRT1 and not SIRT6, so maybe knockdown of SIRT1 would allow them to determine if this observed repression of NF-kB genes is due only to SIRT6 and not some other interaction with SIRT1.
Scientific paper Reference
Mark Devries
Email [email protected]
last updated 1/22/09
http://www.gen677.weebly.com
