This web page was produced as an assignment for Gen677 at UW-Madison Spring 2009
Sumolation search
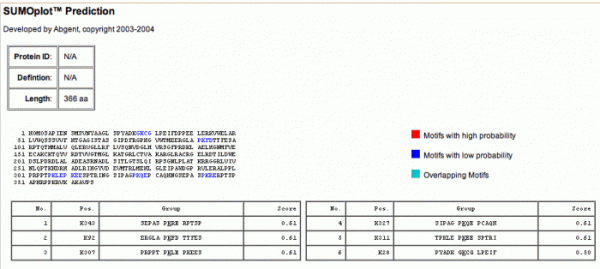
Sumolation is a non covalent modification where a 10KD protein is attached to specific lysine of proteins. Sumolyation differs from ubiquitination in that it dosent actually lead to protein degradation but has been shown to be involved in transcriptional silencing genomic stabilization and the stress response (Johnson, 2004). The reason I choice to look at Sumolyation is because SIRT1, family member of sirtuins, was found to be sumolated. And the fact that Sumolyation modifications are used in DNA damage response maybe me interested in if SIRT6 could also be sumolated. So using the Abgent SUMOplot predictor I was able to come up with different sites of where SIRT6 could possible be sumolated. Most of the motifs that I found that could be sumolated had a low probability of actually being a sumolation site.
Predicted Phosophorlation sites
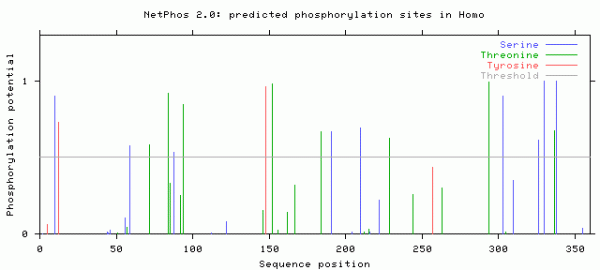
I obtained the predicted phosphorylation sites from NetPhos 2.0 shown here in this graph is the predicted sites of phosphorlation by different kinases. The X axis shows what position that these phosophorlation events occur, while the Y axis shows the probability of these sites being true. The threshold is set at .5 so an phosphorlation site above .5 is considered a most likely a phosphorlation site.
Location of SIRT6
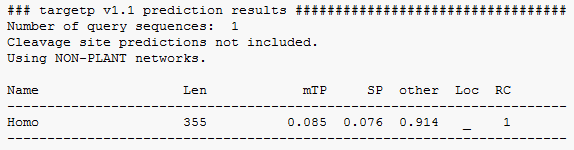
I also used TargetP to see if the prediction of where SIRT6 would locate too was true to the experimentally determined location of SIRT6. So I used the full length FASTA peptide sequence of SIRT6 human and found that the predicted location most probable corresponds to the experiment location of the Nucleus. The reason I say probably is it was considered to locate so place other then the mitochondria or sectory pathway. But other could mean cytoplasm or nucleus so I think it is right.
References
1. http://www.abgent.com/tools/sumoplot?[recipient_id]
2. Johnson, E. (2004). Protein modification by SUMO. Annual rev biochem. 73, p355-382
3. http://www.cbs.dtu.dk/services/NetPhos/
4. http://www.cbs.dtu.dk/services/TargetP/
