Phenotypes
This web page was produced as an assignment for Gen677 at UW-Madison Spring 2009
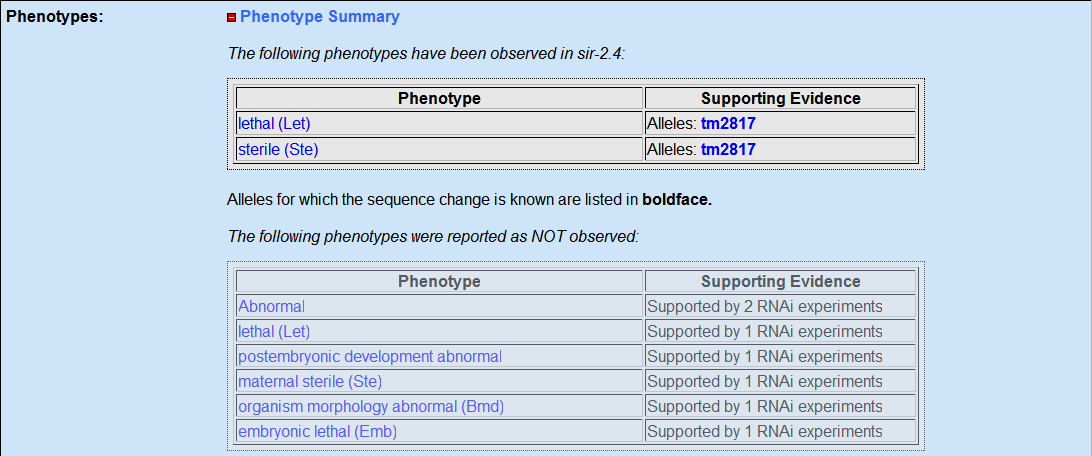
Wormbase results
I went about finding SIRT6 or the C elegans homolog sir-2.4 by performing a search in wormbase, I found out that it is related to the SIR family of proteins. The gene is a predicted gene that was curated and its genomic location in C. elegans is I:5,991,073..5,992,630. The phenotypes for the RNAi experiment showed only wildtype phenotype for the sir-2.4 gene. But interestingly the only the mutant forms of the sir-2.4 gene showed a phenotype that makes the worms sterile and lethal.
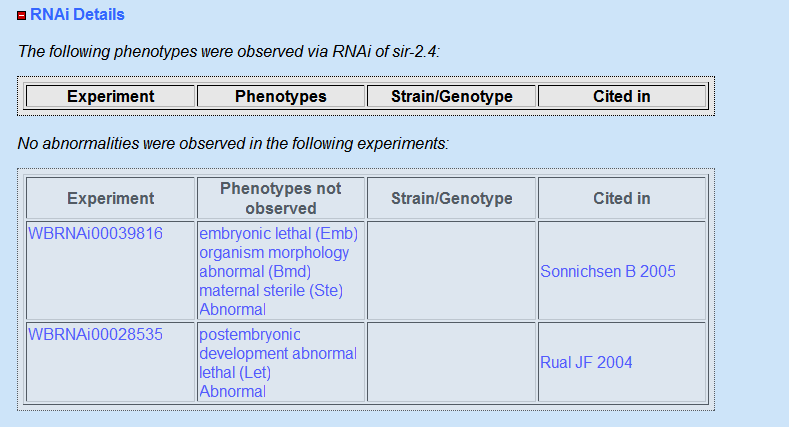
RNAi results
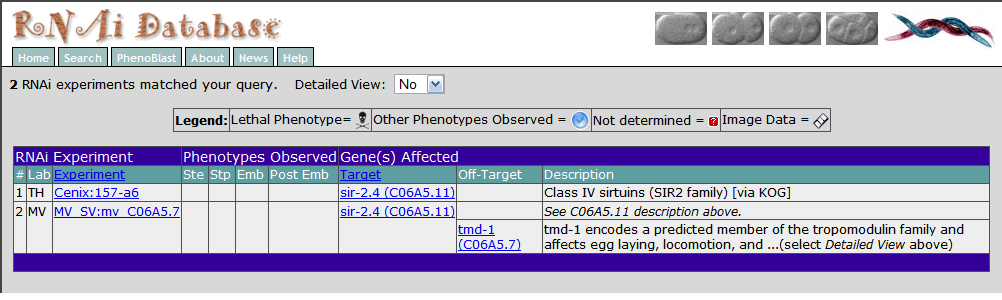
I also looked at sir-2.4 in RNAi data base I again found similar information that there was no phenotype observed from RNAi knockdown of sir-2.4. The experiment that looked at RNAi knockdown of sir-2.4 was called Cenix:157-a6 and they performed RNAi knockdown by injection. So it seems that sir-2.4 does not have an observable phenotype so I went to looking at its close relative sir-2.1 or SIRT1 in humans. The sir-2.1 paralog does have a phenotype which is embryonic lethal.
Conclusions
It was surprising to me that the SIRT6 gene didn't have any phenotypes considering that others have seen phenotypes like Kawahara saw that mice missing SIRT6 would live only to 1 month. So I was expecting at least find a embryonic lethal phenotype, but maybe RNAi does not have the same effect as an animal total missing the SIRT6 gene.
Chemical activators or inhibitors ?
SIRT6 importance has only recently become evident due to this little information is known about what chemicals can activate or inhibit the SIRT6 gene. From studies by Kanfi the only known 'activator' is nutrient availability, this could mean that NAD+ is an activators but no direct study has been done to say for sure that NAD+ does. I would suspect that NAD+ would because it activates other paralogies like SIRT1,2,3. And there has been no studies done yet of what chemicals could inhibit SIRT6. So this is one area of study that could allow for better understand of SIRT6 function by allowing to knock SIRT6 out/activate at different times in a organisms life.
From PubChem retrieved from http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=925
References
1. Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, Cohen HY. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008; 582(5):543-548.
2. www.wormbase.org
3.http://www.worm.mpi-cbg.de/phenobank2/cgi-bin/MenuPage.py
Mark Devries
Email [email protected]
last updated 1/22/09
http://www.gen677.weebly.com